B2: Interaction of biological and computational neuronal networks in a hybrid circuit
Ulrich EgertP, Karl-Heinz BovenE and Martin NawrotQ
Q = Bernstein Center for Computational Neuroscience, Berlin
E = Multi Channel Systems GmbH
P = Biomicrotechnology
Scientific background
Most theories on network function depend on statistically, but not topologically, defined connectivity. Simple, unstructured biological neuronal networks in cell cultures (cBNN) should thus be able to represent 'information', form 'memory', and perform 'computational processing', e.g. of stimulation patterns derived from other networks. Hybrid feedback systems of cBNNs and computational neuronal networks (CNN) have been described, but there is no general concept for a technical interface. cBNN activity cannot yet be controlled in a meaningful way, lacking, in part, a concept of 'meaning'. CNNs, on the other hand, are tools superior to classical control systems when control parameters are insufficiently defined. In vivo and in vitro studies report that BNNs can be in different activity states, e.g. 'up' and 'down'-states, with distinct subthreshold membrane potentials, input resistance and network activity, with some activity patterns in cBNNs resembling oscillations. Likewise, large cortex-like CNNs develop various activity states in terms of synchrony of spiking activity and its temporal pattern.
Objectives
We aim to find out how nervous systems maintain stability of their activity, and simultaneously respond to incoming activity and store information, to understand how recurrent activity patterns in BNNs develop and propagate, how they interact and how this can be dynamically controlled. We will investigate activity dynamics in hybrid systems of connected cBNN and CNN, feeding activity recorded in a BNN to a CNN and stimulating the BNN based on the activity in the CNN. The approach combines the control possible in CNNs with the complexity and plasticity of BNNs.
Associated Postdoc-Project: "Interacting with Biological Neuronal Networks – Contributions of Spontaneous Activity and Neuron-Glia Interaction" (Dr. Matthew Goddard)
Interacting with biological neuronal networks is at the core of numerous research and clinical applications. Be it deep brain stimulation for affective disorders [1,2], 'brain pacemakers' for Parkinson's disease or epilepsy [4-5], or cochlear and retinal implants or the sensory impaired [for a review see 3]. In each case the goal of the interface is the same: to achieve a means of synthetic communication with the nervous system.
Although such technologies have clear benefits to the patient, the effects of stimulation on networks remain far less clear than what their treatment efficacies might predict. This unfortunately means that more complex interactions with networks are in practice quite difficult to achieve effectively. In the simplest sense, interaction can be achieved by artificially depolarising via their axons a small group of neurons with the use of invasive stimulating microelectodes [6-8]. This requires not only that any given stimulus will influence single neurons, but also that the egressing effects spread outward and influence the activity dynamics of the entire system or subsystem. Notwithstanding the technical limitations of neuronal-machine interfaces, the extent to which network stimulation will be successful will depend on how well the dynamics of the system can be defined and translated into stimulus paradigms.
Taking advantage of known principles thereof - namely connectivity pathways, individual neuronal properties, synaptic plasticity, and the state of the network itself [9-12] - it should be possible to achieve more complex interactions than those currently in clinical use such as the ability to artificially stimulate a network in such a way that it 'remembers' stimulus paradigms, and even to predict certain response properties for given inputs. However, much of the variability in network responses has yet to be attributed to underlying governing principles, which is why many current models can only offer low predictability. In clinical practice, attempts to translate patterns of electrical activity during epileptic seizures into pacemaker stimulation currents is proving to be highly challenging [12-14].
This is because the status of the network parameters are essentially unknown and so typical stimulation parameters must be adjusted iteratively by trial and error. As a result, the outcome of such stimulations are highly variable and may even be ineffective at times. One commonly overlooked principle in neural-engineering is the contribution that glia have on network dynamics; despite the fact that is now known that glia can influence network synchronization through a variety of mechanisms related to modulation of the extracellular fluid environment [15-19]. Thus, what this project aims to achieve is a more detailed understanding of neuron-glia intereactions during spontaneously active and stimulated network states. The results of this project will contribute to the improvement of neurotechnical interfaces, to our understanding of how local activity spreads into neuronal networks and, generally, help us to improve the predictably and reproducibly induce a desired state of activity in a highly dynamic neuronal environment
 |
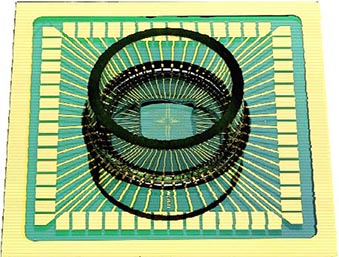 |
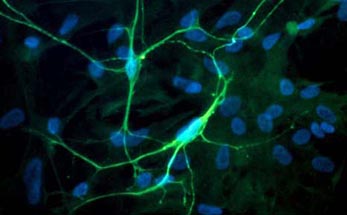
Fig. 1 Left: Primary culture of neurons from embryonic spinal cordstained for neurofilament. Nuclei stained for DNA stain (blue) shows the non-neuronal cell, which are mostly astrocytes. Right: Microelectrode Array (MEA). The recording electrodes are at the ends of the substrate-integrated leads, forming a grid of 60 electrodes in the center of the carrier slide (outer dimensions 5 x 5 cm).
 |
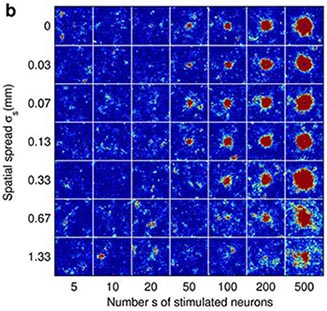 |
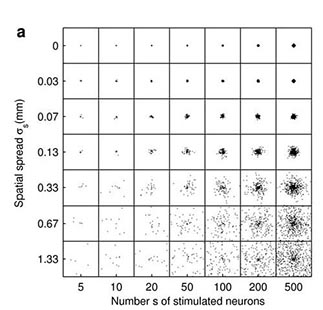
Fig 2. Cortical network model (2 mm x 2 mm) response to a synchronous stimulus of varied strength and spatial spread. (a) Locations of stimulated neurons indicated by black dots (b) Network response; color coding shows the number of spikes from the excitatory populationfrom 1-2 ms, i.e. around one synaptic delay (1.5 ms) after the stimulus. (From Mehring et. al (2003) Biol. Cybern).

